|
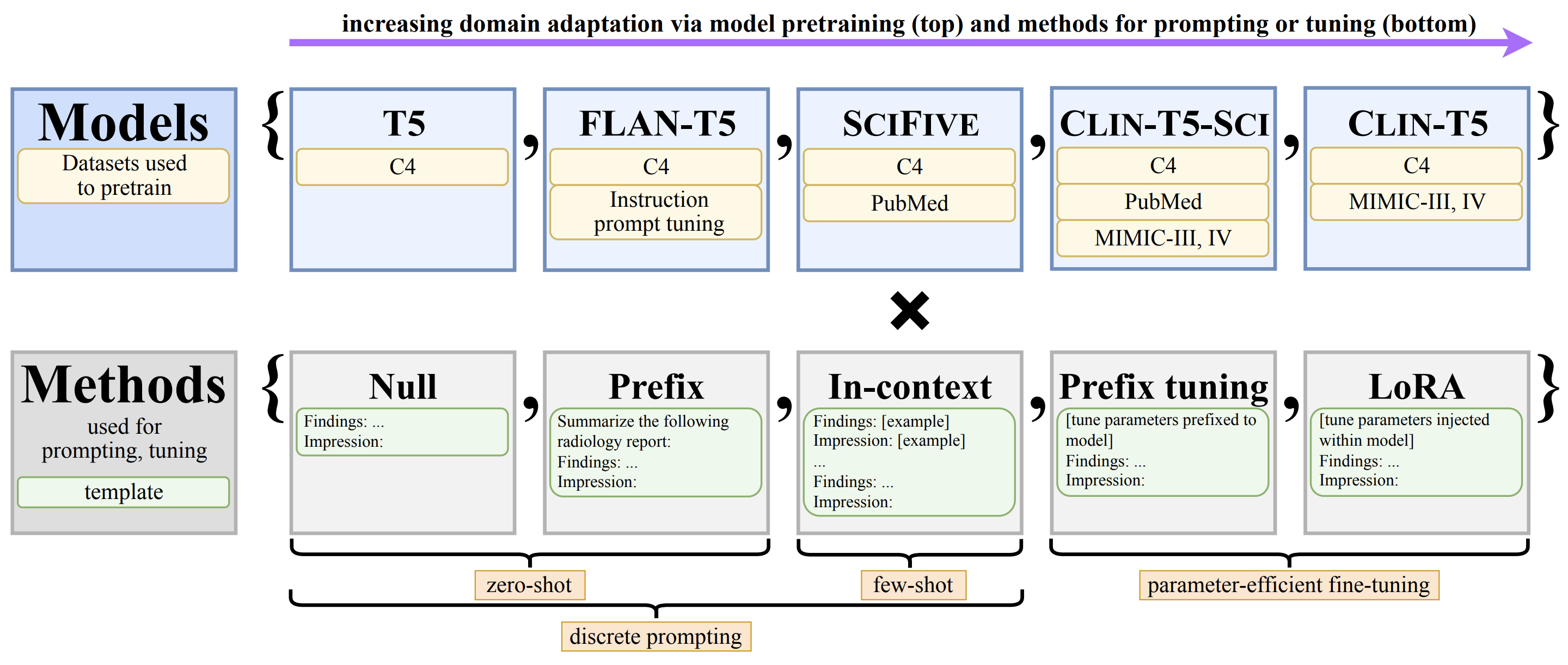
We systematically investigate lightweight strategies to adapt large language models (LLMs) for the task of radiology report summarization (RRS). Specifically, we focus on domain adaptation via pretraining (on natural language, biomedical text, or clinical text) and via discrete prompting or parameter-efficient fine-tuning.
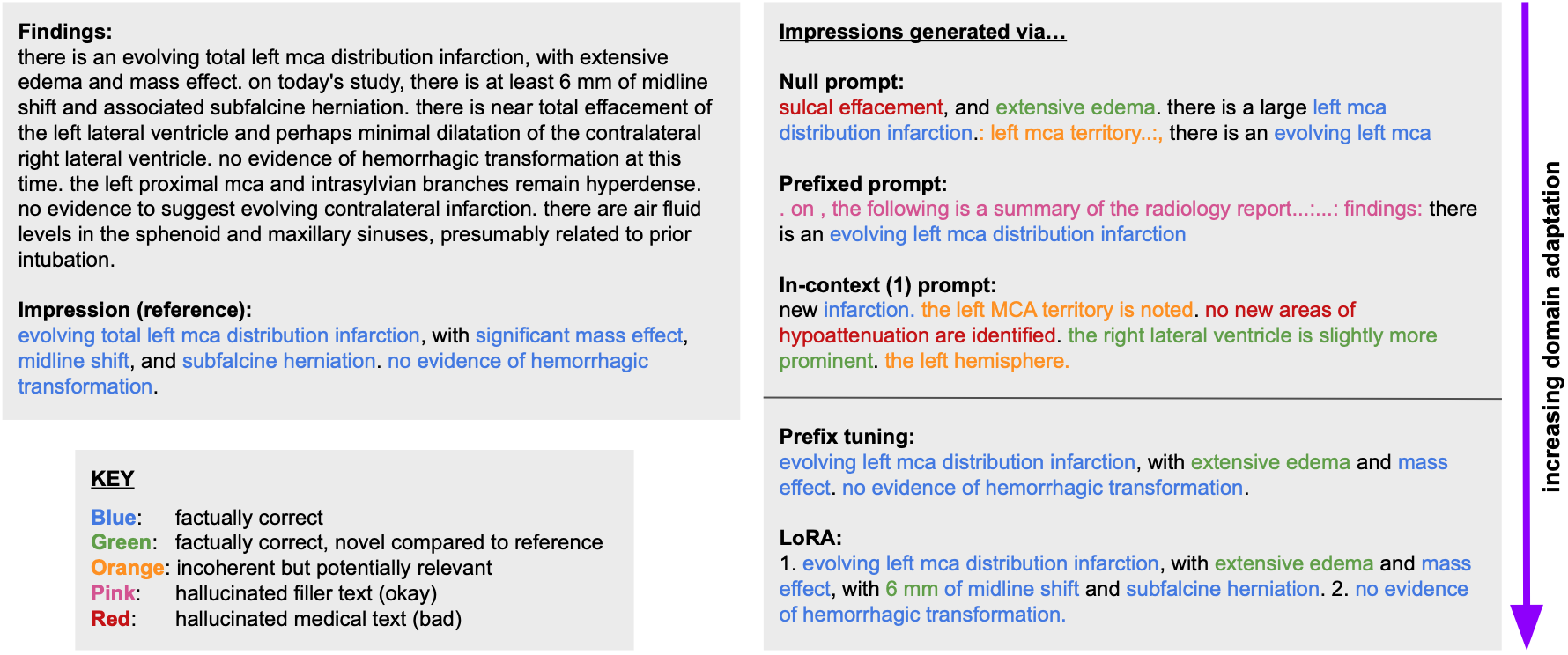
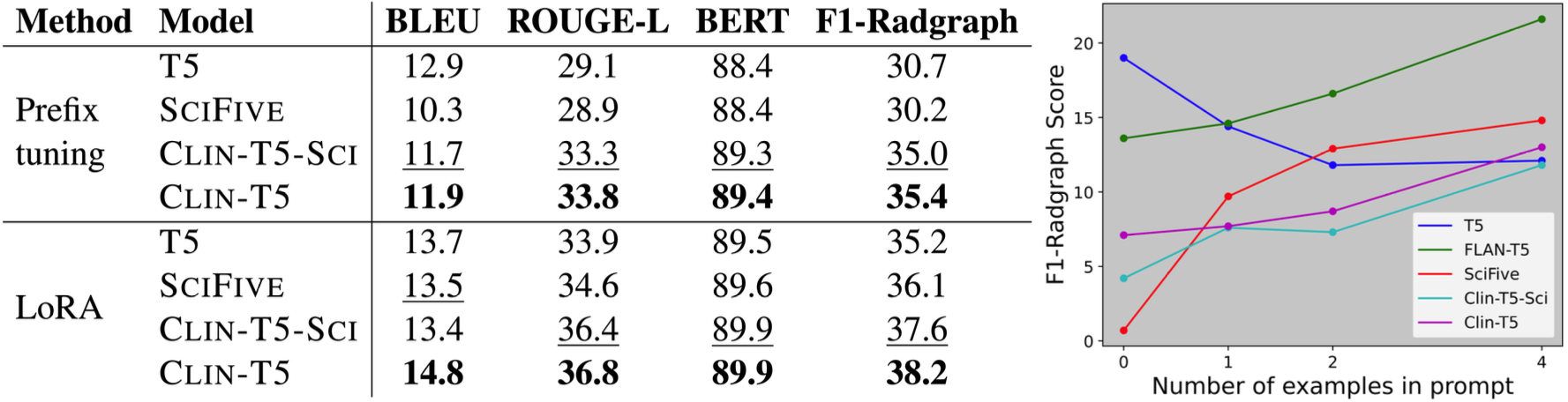
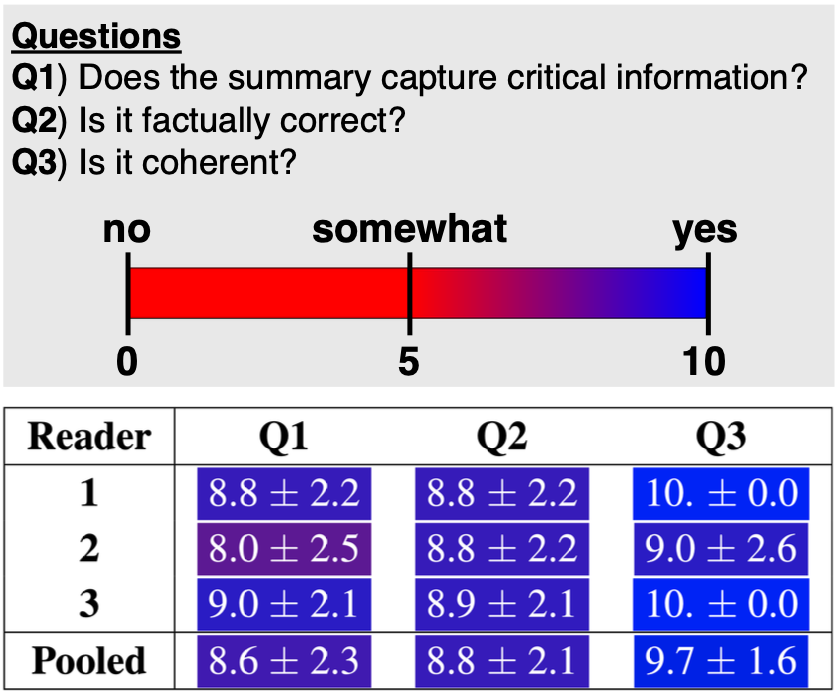
Our results consistently achieve best performance by maximally adapting to the task via pretraining on clinical text and fine-tuning on RRS examples. Importantly, this method fine-tunes a mere 0.32% of parameters throughout the model, in contrast to end-to-end fine-tuning (100% of parameters). Additionally, we study the effect of in-context examples and out-of-distribution (OOD) training before concluding with a radiologist reader study and qualitative analysis.
This research highlights the importance of domain adaptation in RRS and provides valuable insights toward developing effective natural language processing solutions for clinical tasks.
|